Alternative Fitting Procedure with Surrogate Loss Function¶
This example demonstrates an alternative way to fit a boosted model using a for loop such that holdout loss functions amongst other things can be customized.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.datasets import make_classification
from sklearn.preprocessing import scale
from sklearn.model_selection import train_test_split
from sklearn.metrics import roc_auc_score
from genestboost import BoostedLinearModel
from genestboost.weak_learners import SimplePLS
from genestboost.link_functions import LogitLink
from genestboost.loss_functions import LogLoss
%matplotlib inline
Create a Dummy Classification Dataset¶
X, y = make_classification(n_samples=20000,
n_features=50,
n_informative=20,
weights=(0.85, 0.15),
random_state=11,
shuffle=False)
X = scale(X)
Alternative Fitting Procedure¶
X_train, X_val, y_train, y_val = (
train_test_split(X, y, test_size=0.30, stratify=y, random_state=13)
)
# notice no validation set arguments in the init - we will compute holdout in our loop below
model = BoostedLinearModel(
link=LogitLink(),
loss=LogLoss(),
model_callback=SimplePLS, # for now, still need to specify this arg
model_callback_kwargs={},
alpha=5.0,
step_type="decaying",
weights="newton",)
# HELPER
def calc_roc(yp, yp_val):
"""Closure of y_train and y_val."""
return (roc_auc_score(y_train, yp), roc_auc_score(y_val, yp_val))
# instead of using fit, we will use a for-loop to fit the model while using
# ROC-AUC on the holdout set to determine stoppage
yp, eta_p = model.initialize_model(X_train, y_train) ### IMPORTANT - initializes the model
eta_p_val = model.decision_function(X_val)
yp_val = model.compute_link(eta_p_val, inverse=True)
loss_list = [calc_roc(yp, yp_val)] # rocauc loss [(train, val)]
# main loop
max_iterations, min_iterations, iter_stop = 2000, 20, 20
for i in range(max_iterations):
yp, eta_p = model.boost(X_train, y_train, yp, eta_p, SimplePLS, {})
eta_p_val += model.decision_function_single(X_val) # predict on only the last model for performance
yp_val = model.compute_link(eta_p_val, inverse=True)
loss_list.append(calc_roc(yp, yp_val))
if i >= min_iterations and i > iter_stop:
loss_check = loss_list[-iter_stop][1]
if loss_list[-1][1] < loss_check:
break
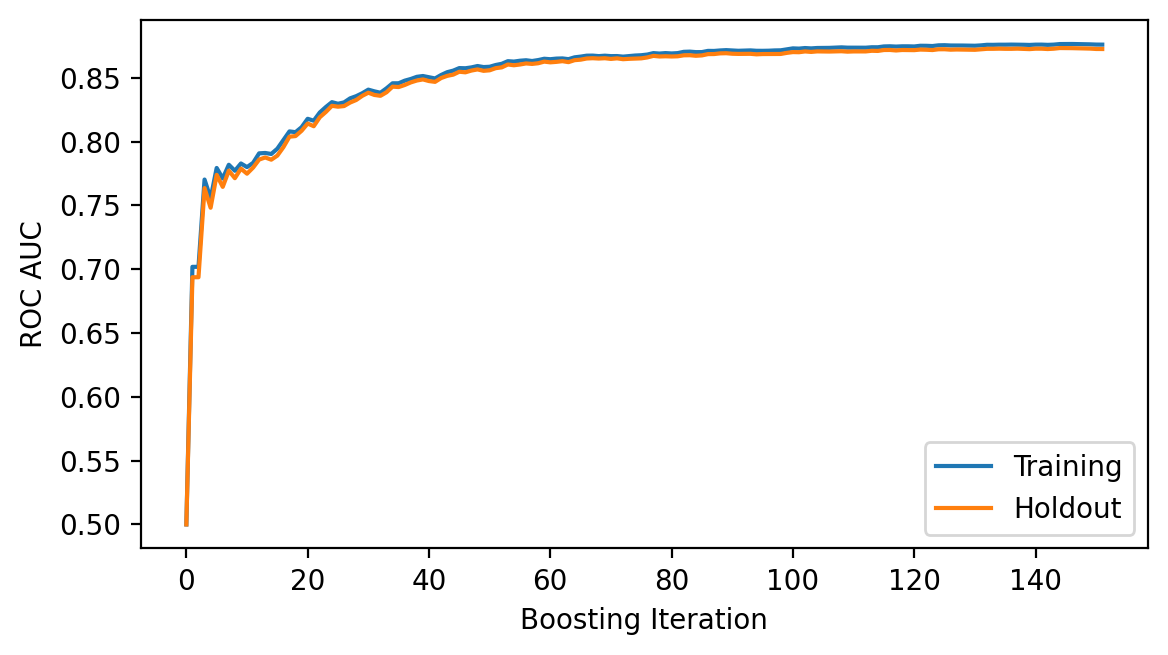
print("Number of Boosting Iterations: {:d}".format(model.get_iterations()))
Number of Boosting Iterations: 151
Plot the loss history¶
fig = plt.figure(figsize=(6.5, 3.5), dpi=200)
ax = fig.add_subplot(111)
ax.plot(np.array(loss_list), label=["Training", "Holdout"])
ax.legend(loc="best")
ax.set_ylabel("ROC AUC")
ax.set_xlabel("Boosting Iteration");
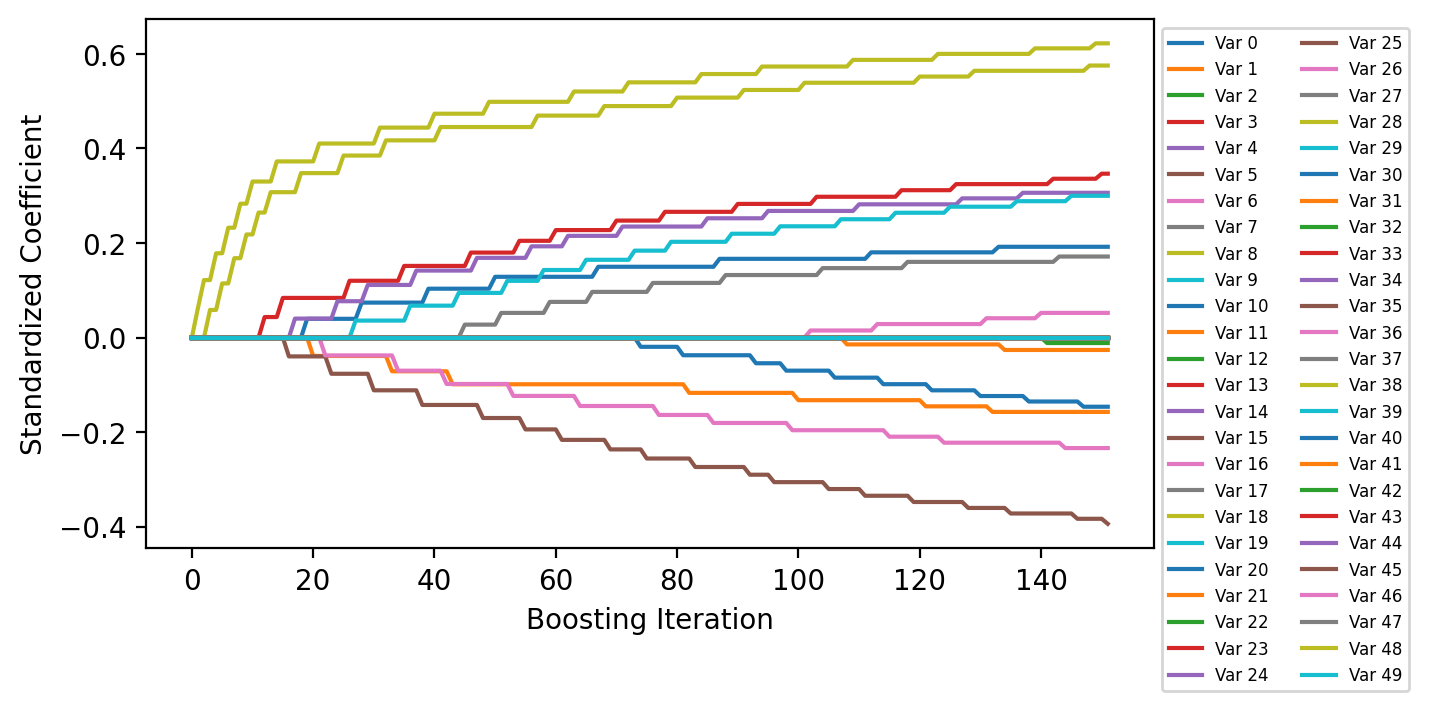
Plot Coefficient History¶
The coefficients are scaled by the standard deviation of the corresponding features in the data set to get standardized coefficients.
fig = plt.figure(figsize=(6.5, 3.5), dpi=200)
ax = fig.add_subplot(111)
ax.plot(model.get_coefficient_history(scale=X.std(ddof=1, axis=0)), label=[f"Var {i:d}" for i in range(X.shape[1])])
ax.legend(loc="upper left", bbox_to_anchor=(1, 1), ncol=2, fontsize=6)
ax.set_xlabel("Boosting Iteration")
ax.set_ylabel("Standardized Coefficient");
Order that Variables Entered the Model¶
print("Number of Selected Variables in the Model: {:d}".format(len(model.get_coefficient_order())))
model.get_coefficient_order()
Number of Selected Variables in the Model: 14
[8, 18, 3, 5, 14, 0, 1, 6, 19, 17, 10, 16, 11, 2]